Glycan Profiling
In Depth Structure Analysis and Enhanced Quantification of Glycans via UPLC-FLR-MS
The N-glycosylation represents an important and equally variable posttranslational modification of biotherapeutics. Generally requested as a release criteria by authorities, it is important to deliver thorough and reliable analysis of the N-glycans of your protein.
The composition of N-glycans may change the stability, efficacy, or bioavailability of your product. Since slight changes in the production process may alter the N-glycosylation, monitoring product glycosylation throughout process development is inevitable.
What are the most abundant N-glycans of mammalian cells?
Glycostructures on mammalian proteins and cells are built like pearl chains from monosaccharide components such as sialic acid (N-acetylneuraminic acid or N-glycolylneuraminic acid), mannose, galactose, fucose, N-acetylglucosamine (bisGlcNAc) and N-acetylgalactosamine.
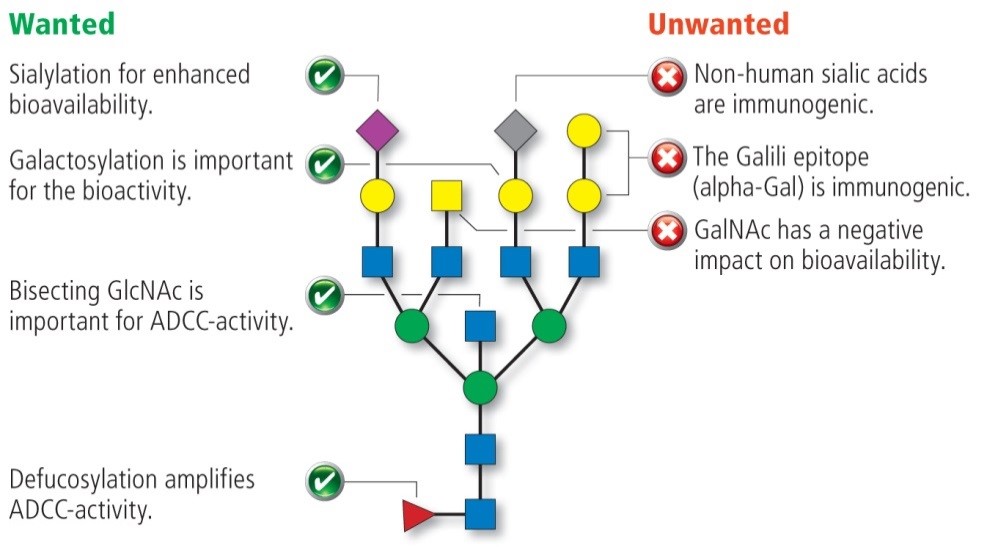
The capping of the N-glycans with sialic acids is one of the most important characteristic, since it enables ionic interactions and produces charge variants of glycoproteins. In humans, it is found in 2,3- and 2,6-linkage while another linkage variant is known to be immunogenic. N-glycolylneuraminic is observed in therapeutics produced in rodent cells.
A high degree of galactose on the N-glycan´s antennae improves the bioactivity of the corresponding proteins, whereas the presence of N-acetylgalactosamine (GalNAc) has a negative effect on the bioactivity
The absence of core fucose has often been described to boost the ADCC activity of monoclonal antibodies (mAbs). Defucosylation may also lead to an increase in bisecting N-acetylglucosamine (bisGlcNAc) in many proteins although its correlation with ADCC activity of mAbs remains unclear.
How is the analysis of N-glycans performed?
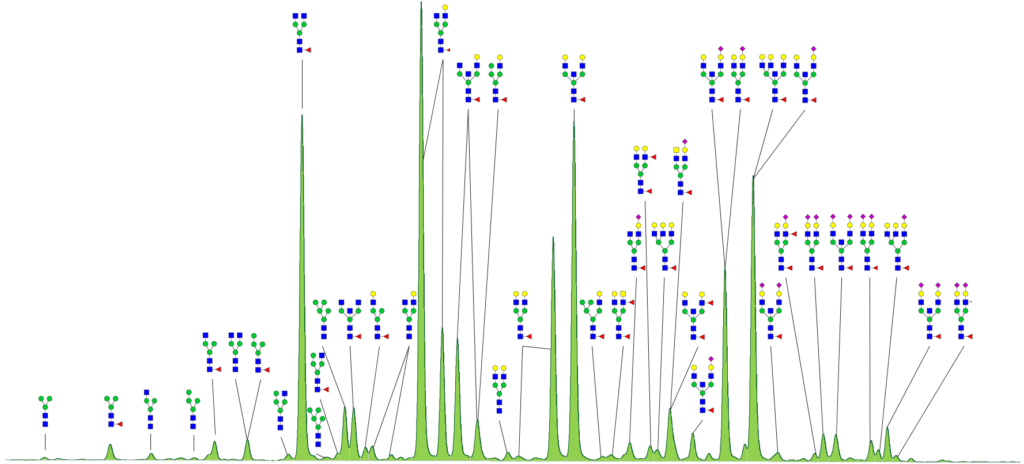
N-glycan analysis starts with releasing glycans form the glycoproteins. Efficient deglycosylation is achieved through digestion with specific N-glycosidase F. The released N-glycans are labeled with a fluorescent dye and analyzed via high resolution HILIC-UPLC-FLR. HILIC-UPLC provides a superior resolution allowing to easily recognize slight changes in N-glycosylation profile.
Furthermore, the fluorescence method is quantitative, since every N-glycan is labeled with only one fluorescent dye molecule. Fractions of the different glycan species are obtained from integration of the corresponding peak area.
Due to the stereoselectivity of the HILIC phase N-glycans of identical masses can be differentiated. For example, galactosylation on the 3-arm or 6-arm and 2,3- or 2,6- linkage of sialic acids can be distinguished by their retention time.
Coupling with ESI-QTOF-CID-MS/MS devices allows reliable identification of N-glycan structures on base of characteristic fragment spectra and matching to a spectra library. Thereby, a further differentiation of isobaric N-glycans, comprising bisecting GlcNAc, antennary GlcNAc or unwanted GalNAc is also feasible.
In contrast to the fluorescence method, mass spectrometry requires multiple charge states and different numbers of isotopes to regard for a quantitative approach. Even then the same ionization efficiency cannot be guaranteed. MS quantification can reasonably be applied only for physicochemically similar N-glycans.
With our approach we employ both methods, fluorescence and mass spectrometry, and combine the individual advantages for N-glycosylation analysis. GlycoFiler® offers a reliable and fast quantification and calculation of biologically relevant glycosylation parameters. Each sample will be compiled in a certificate of analysis (CoA) comprising the fluorescence chromatogram, information about the sample and the applied method, the measured and calculated N-glycosylation parameters of biological relevance like sialylation, fucosylation, high mannose content, antennarity and more. For validation purpose the performance of a running standard is monitored and included in the CoA.
Thanks to automated evaluation, first results can be obtained immediately after measurements. The required sample amount for a typical GlycoFiler® workflow is only 15 µg of glycoprotein.
Besides N-glycan profiling we also perform the analysis of O-glycosylation. This is done by chemical release of O-glycans and quantification of fluorescence peaks after labeling with 2-AB tag.
Do you need more information about our N-glycoprofiling services?
Frequently Asked Questions (FAQ) for Glycan Profiling Services
Glycans contribute significantly to the heterogeneity of the protein’s mass and charge and can also impact protein activity. During development glycan profiling can be used to (i) identify the best clone, (ii) the best media and/or (iii) the best process. Additionally, during clinical manufacturing and market supply glycan profiling can be used to monitor product quality. In later stage clinical trials and market supply glycan profiling is often used as release test.
Of course, there have been various methods, but we believe the most suitable technique is the enzymatic release of N-glycans and fluorescent labeling. Glycans are separated and quantified via HILIC chromatography and identified by MS/MS. In case of overlapping peaks, the peak area is hybridized with MS signal intensities to give you the most accurate relative quantification for each glycan structure. We have developed GlycoFiler®, a software solution for reliable and fast quantification of N-glycan profiles.
N-glycosylation of asparagine can occur if the amino acid sequence comprises the consensus sequence NXS/T (where X is any amino acid except for P). O-glycans are detected on serine or threonine residues, but also on hydroxylysine or hydroxyproline.
The number of different glycosylation sites is most easily determined via LC-MS/MS peptide mapping using proteolytic digests. Glycosylated peptides are identified from the sequence information and correlation to characteristic MS fragments of glycan structures.
For N-glycan profiling a minimum of 15 µg protein is required.
For more relevant information and additional FAQs check also the FAQ section in our resource center.
Glycan Profiling Services at FyoniBio – Related Content
Read about how our GlycoFiler® software can be used for automated glycan identification in mass spectrometric data analysis.